paired end sequencing read length
We have previously shown how different enrichment methods perform with respect to covered regions underrepresented regions and sequencing efficiency. As long as you dont exceed the maximum number of cycles you will be fine.

Sequencing Read Length How To Calculate Ngs Read Length
B no overlap between the paired-end.

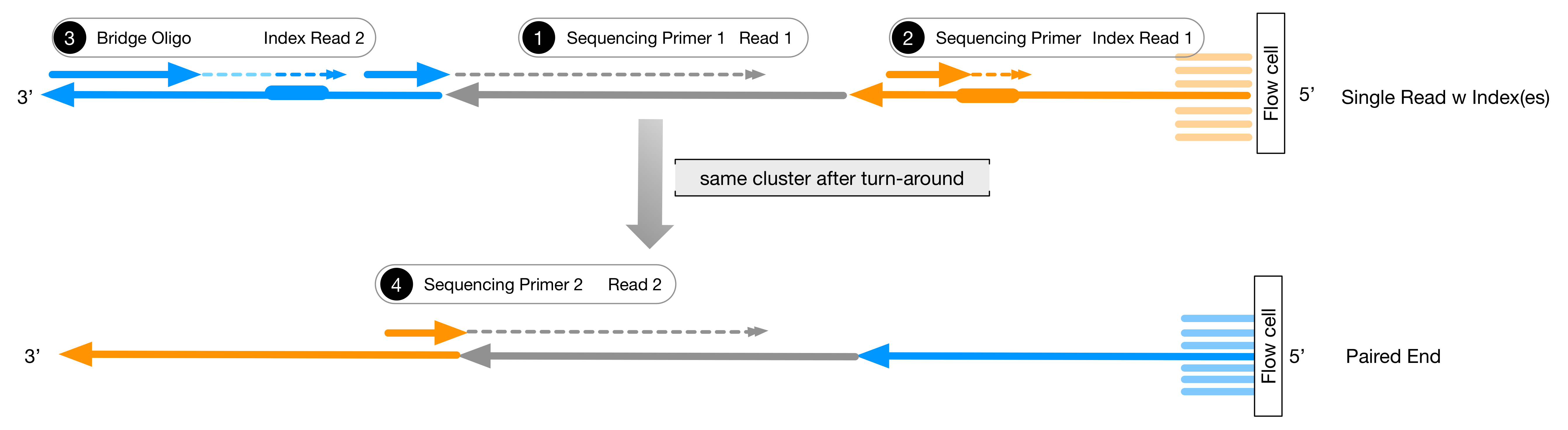
. Paired-end sequencing allows users to sequence both ends of a fragment and generate high-quality alignable sequence data. Refer to the How many cycles of. To ensure the highest level of quality Illumina supports reads up to a certain length depending on the sequencing platform and SBS kit version.
For Illumina kits for example you include R1 and R2 length in the sample sheet. In this Tech Note we focus on. Paired-end 150 means that one read of 150 bases in size is generated from each end of the fragment through the inserted middle piece of target DNA from both directions for a total of 2.
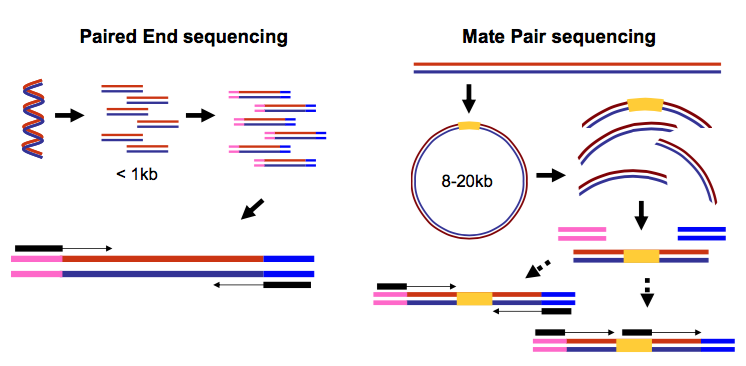
The library prep protocols are designed to. Whole genome sequence construction is becoming increasingly feasible because of advances in next generation sequencing NGS. The paired-end short read lengths are always 2 x 150bp 300bp.
In paired-end reading it starts at one read finishes this direction at the specified read length and then starts another round of reading from the opposite end of the fragment. Three possible scenarios for paired-end read lengths and target DNA fragment lengths. The Illumina paired-end sequencing technology can generate reads from both ends of target DNA fragments which can subsequently be merged to increase the.
A Short overlap between the paired-end reads. We use an Illumina MiniSeq for our short-read sequencing runs. The depth of the sequencing coverage the length of.
There are several factors to consider when planning a next generation sequencing NGS experiment. Paired-end sequencing facilitates detection of genomic. Why does paired-end sequencing help assembly.
For a 150 cycle kit.

Choosing The Right Read Length For Diagnostic Sequencing Cegat Gmbh
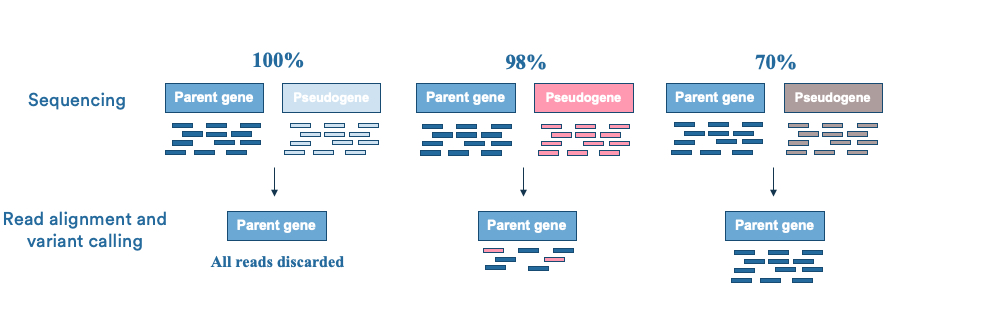
Blueprint Genetics Approach To Pseudogenes And Other Duplicated Genomic Regions Blueprint Genetics
What Is Paired End 150 Omega Bioservices
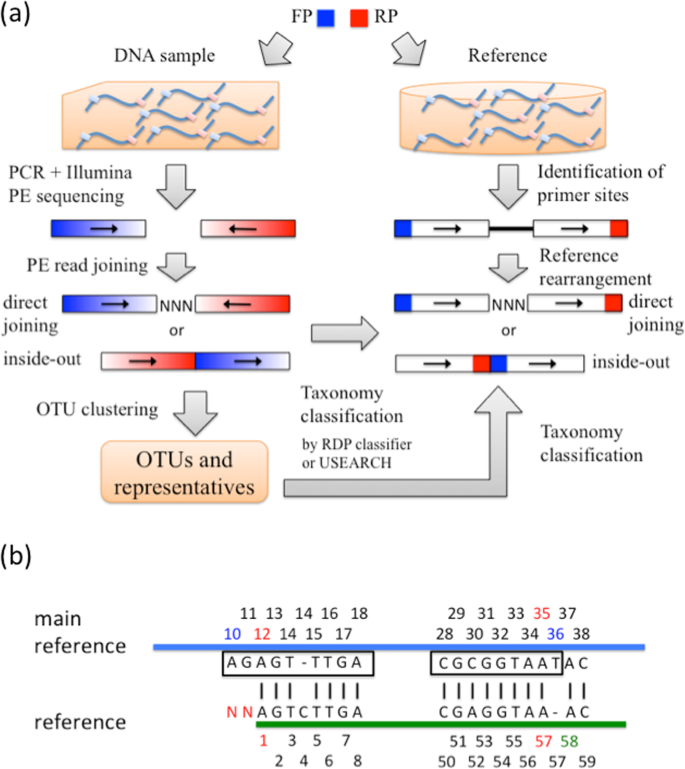
Joining Illumina Paired End Reads For Classifying Phylogenetic Marker Sequences Bmc Bioinformatics Full Text
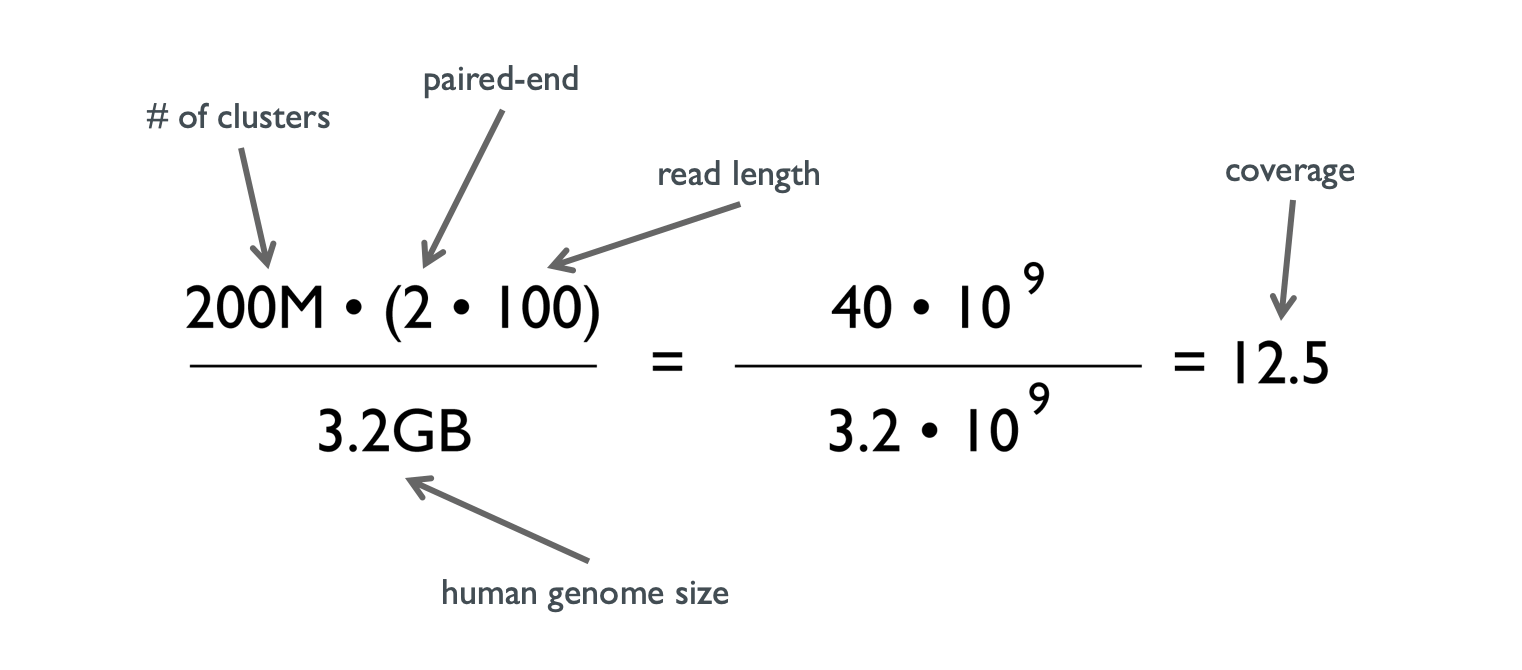
How To Calculate The Coverage For A Ngs Experiment

Illumina Sequencing Illumina Sequencing By Synthesis 1010genome Quality Ngs Bioinformatics Data Analysis Services
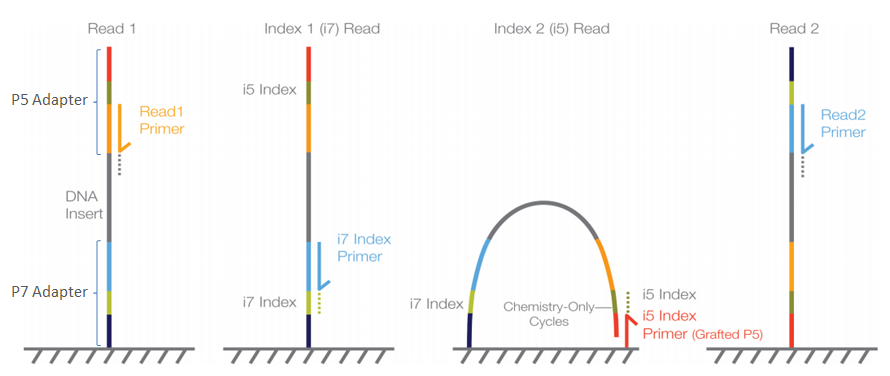
Adapter Trimming Why Are Adapter Sequences Trimmed From Only The 3a Ends Of Reads

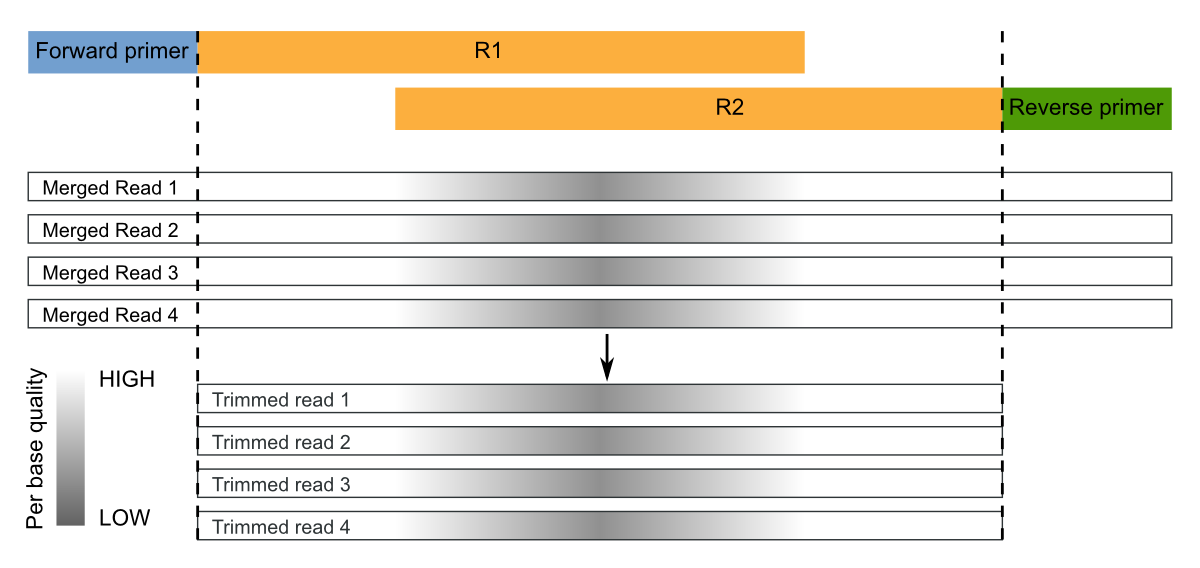
Effect Of Truncation Length Of Sequencing Reads On Merging Paired End Download Scientific Diagram
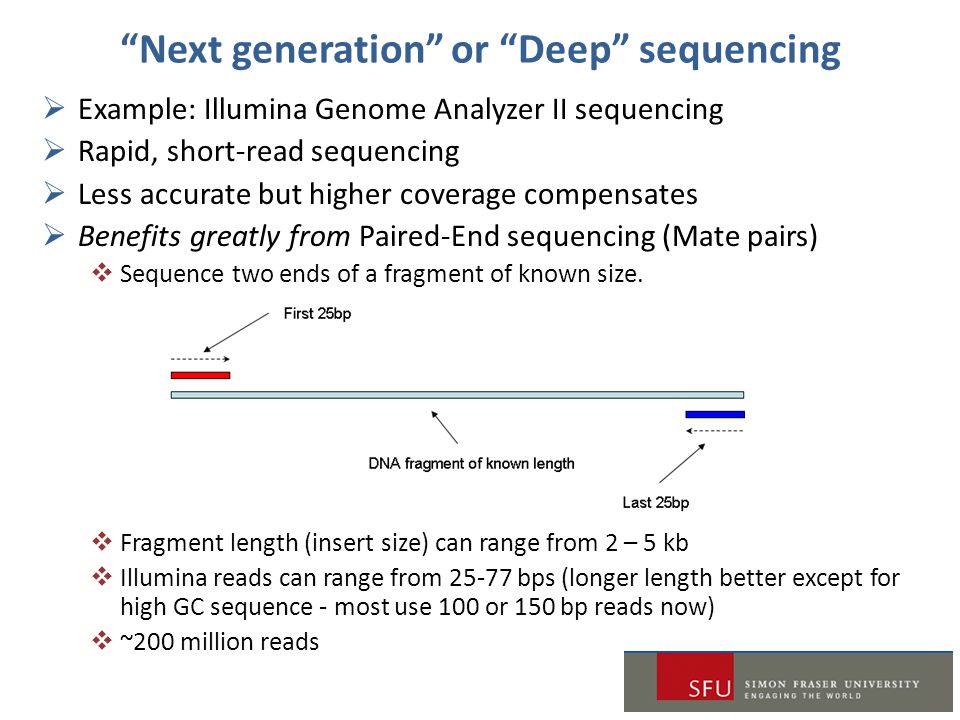
Ngs Bioinformatics Workshop 2 Ppt Download
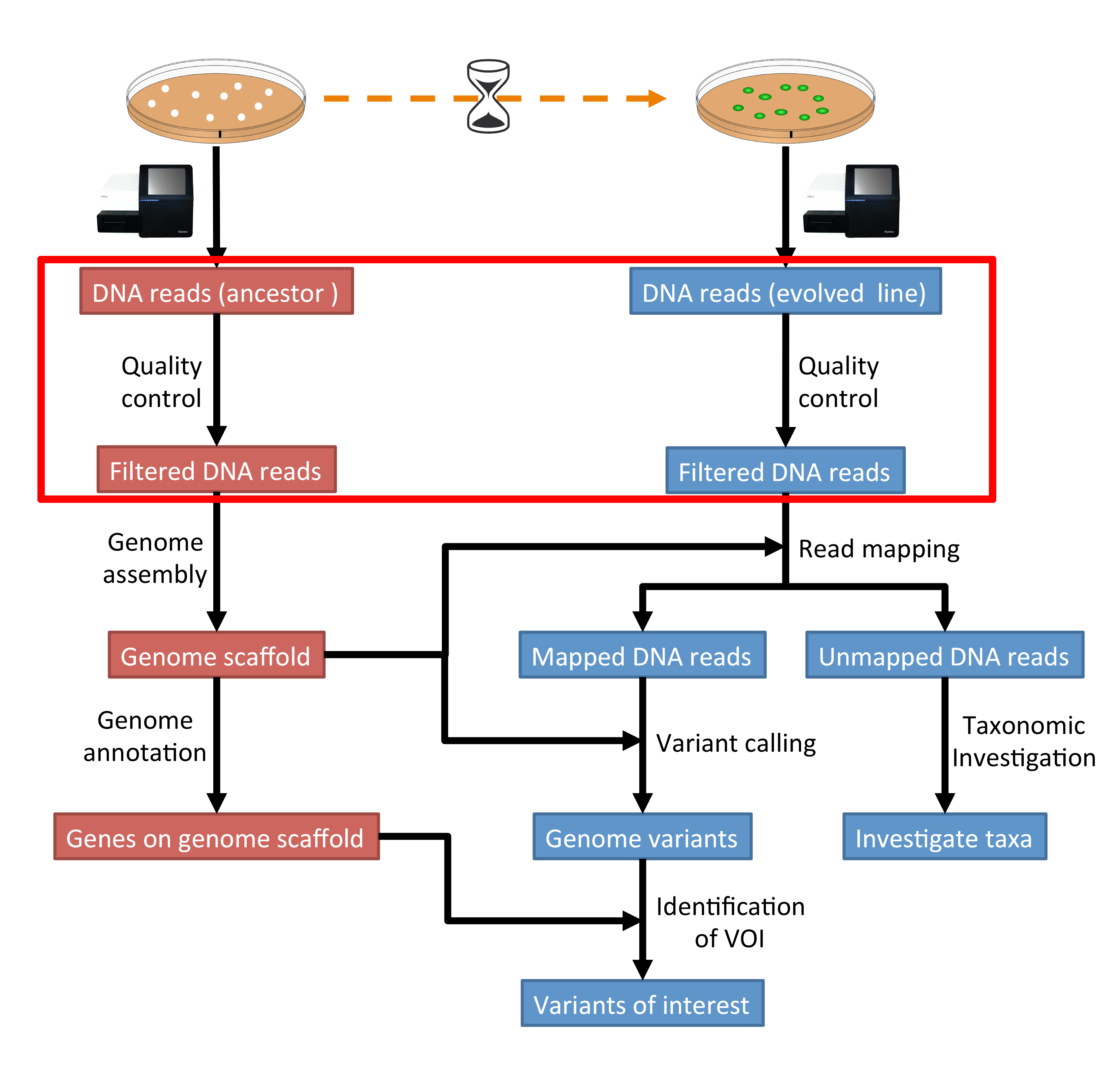
3 Quality Control Genomics Tutorial 2020 2 0 Documentation

Paired End Sequencing Left Showing Read 1 And Read 2 Primers Starting Download Scientific Diagram
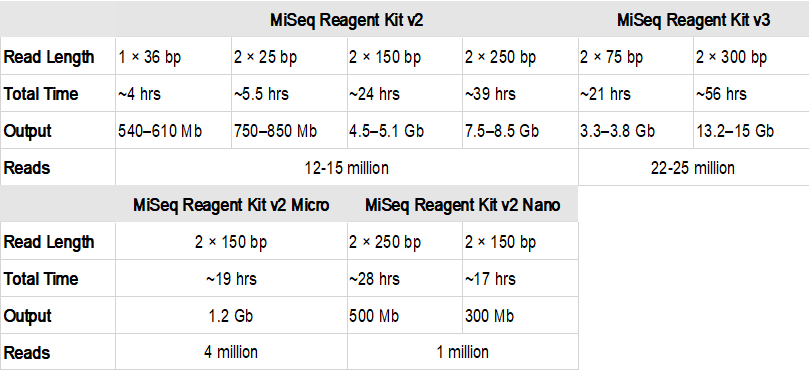
Short Read Sequencing Genomics Core Ecu

Illustrations Of Paired End Sequencing A Illustrates Two Strands Of Download Scientific Diagram
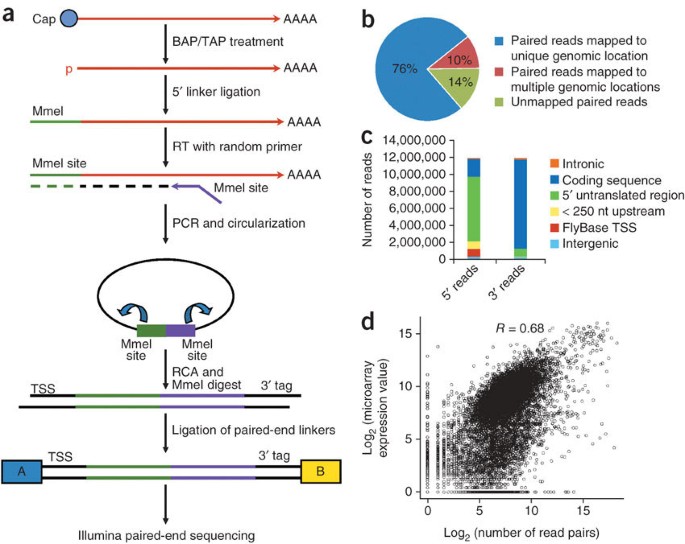
A Paired End Sequencing Strategy To Map The Complex Landscape Of Transcription Initiation Nature Methods